International Chemical Identifier
| Developer(s) | InChI Trust |
|---|---|
| Initial release | April 15, 2005[1][2] |
| Stable release | 1.07.1
/ August 9, 2024 |
| Repository | |
| Operating system | Microsoft Windows and Unix-like |
| Platform | IA-32 and x86-64 |
| Available in | English |
| License | MIT License |
| Website | www |
The International Chemical Identifier (InChI, pronounced /ˈɪntʃiː/ IN-chee)[3] is a textual identifier for chemical substances, designed to provide a standard way to encode molecular information and to facilitate the search for such information in databases and on the web. Initially developed by the International Union of Pure and Applied Chemistry (IUPAC) and National Institute of Standards and Technology (NIST) from 2000 to 2005, the format and algorithms are non-proprietary. Since May 2009, it has been developed by the InChI Trust, a nonprofit charity from the United Kingdom which works to implement and promote the use of InChI.[4]
The identifiers describe chemical substances in terms of layers of information — the atoms and their bond connectivity, tautomeric information, isotope information, stereochemistry, and electronic charge information.[5] Not all layers have to be provided; for instance, the tautomer layer can be omitted if that type of information is not relevant to the particular application. The InChI algorithm converts input structural information into a unique InChI identifier in a three-step process: normalization (to remove redundant information), canonicalization (to generate a unique number label for each atom), and serialization (to give a string of characters).
InChIs differ from the widely used CAS registry numbers in three respects: firstly, they are freely usable and non-proprietary; secondly, they can be computed from structural information and do not have to be assigned by some organization; and thirdly, most of the information in an InChI is human readable (with practice). InChIs can thus be seen as akin to a general and extremely formalized version of IUPAC names. They can express more information than the simpler SMILES notation and, in contrast to SMILES strings, every structure has a unique InChI string, which is important in database applications. Information about the 3-dimensional coordinates of atoms is not represented in InChI; for this purpose a format such as PDB can be used.
The InChIKey, sometimes referred to as a hashed InChI, is a fixed length (27 character) condensed digital representation of the InChI that is not human-understandable. The InChIKey specification was released in September 2007 in order to facilitate web searches for chemical compounds, since these were problematic with the full-length InChI.[6] Unlike the InChI, the InChIKey is not unique: though collisions are expected to be extremely rare, there are known collisions.[7]
In January 2009 the 1.02 version of the InChI software was released. This provided a means to generate so called standard InChI, which does not allow for user selectable options in dealing with the stereochemistry and tautomeric layers of the InChI string. The standard InChIKey is then the hashed version of the standard InChI string. The standard InChI will simplify comparison of InChI strings and keys generated by different groups, and subsequently accessed via diverse sources such as databases and web resources.
The continuing development of the standard has been supported since 2010 by the not-for-profit InChI Trust, of which IUPAC is a member. Version 1.06 and was released in December 2020.[8] Prior to 1.04, the software was freely available under the open-source LGPL license.[9] Versions 1.05 and 1.06 used a custom license called IUPAC-InChI Trust License.[10]
The current software version is 1.07.1 (August 2024), uses the MIT license, and may be downloaded from the InChI GitHub site.
Generation
[edit]In order to avoid generating different InChIs for tautomeric structures, before generating the InChI, an input chemical structure is normalized to reduce it to its so-called core parent structure. This may involve changing bond orders, rearranging formal charges and possibly adding and removing protons. Different input structures may give the same result; for example, acetic acid and acetate would both give the same core parent structure, that of acetic acid. A core parent structure may be disconnected, consisting of more than one component, in which case the sublayers in the InChI usually consist of sublayers for each component, separated by semicolons (periods for the chemical formula sublayer). One way this can happen is that all metal atoms are disconnected during normalization; so, for example, the InChI for tetraethyllead will have five components, one for lead and four for the ethyl groups.[5]
The first, main, layer of the InChI refers to this core parent structure, giving its chemical formula, non-hydrogen connectivity without bond order (/c sublayer) and
hydrogen connectivity (/h sublayer.) The /q portion of the charge layer gives its charge, and the /p portion of the charge layer tells how many protons (hydrogen ions) must be added to or removed from it to regenerate the original structure. If present, the stereochemical layer, with sublayers b, /t, /m and /s, gives stereochemical information, and
the isotopic layer /i (which may contain sublayers /h, /b, /t, /m and /s) gives isotopic information. These are the only layers which can occur in a standard InChI.[5]
If the user wants to specify an exact tautomer, a fixed hydrogen layer /f can be appended, which may contain various additional sublayers; this cannot be done in standard InChI though, so different tautomers will have the same standard InChI (for example, alanine will give the same standard InChI whether input in a neutral or a zwitterionic form.)
Finally, a nonstandard reconnected /r layer can be added, which effectively gives a new InChI generated without breaking bonds to metal atoms. This may contain various sublayers, including /f.[5]
Format and layers
[edit]| Internet media type |
chemical/x-inchi |
|---|---|
| Type of format | chemical file format |
Every InChI starts with the string "InChI=" followed by the version number, currently 1. If the InChI is standard, this is followed by the letter S for standard InChIs, which is a fully standardized InChI flavor maintaining the same level of attention to structure details and the same conventions for drawing perception. The remaining information is structured as a sequence of layers and sub-layers, with each layer providing one specific type of information. The layers and sub-layers are separated by the delimiter "/" and start with a characteristic prefix letter (except for the chemical formula sub-layer of the main layer). The six layers with important sublayers are:
- Main layer (always present)
- Chemical formula (no prefix). This is the only sublayer that must occur in every InChI. Numbers used throughout the InChI are given in the formula's element order excluding hydrogen atoms. For example, "/C10H16N5O13P3" implies that atoms numbered 1–10 are carbons, 11–15 are nitrogens, 16–28 are oxygens, and 29–31 are phosphorus.[11]
- Atom connections (prefix: "
c"). The atoms in the chemical formula (except for hydrogens) are numbered in sequence; this sublayer describes which atoms are connected by bonds to which other ones. The type of those bonds is later specified in the stereochemical layer prefixed by "b". - Hydrogen atoms (prefix: "
h"). Describes how many hydrogen atoms are connected to each of the other atoms.
- Charge layer
- charge sublayer (prefix: "
q") - proton sublayer (prefix: "
p" for "protons")
- charge sublayer (prefix: "
- Stereochemical layer
- Isotopic layer (prefix: "
i"), may include sublayers:[11]- sublayer "
h" for isotopic hydrogen - sublayers "
b", "t", "m", "s" for isotopic stereochemistry
- sublayer "
- Fixed-H layer (prefix: "
f") for tautomeric hydrogens; contains some or all of the above types of layers except atom connections; may end with "o" sublayer; never included in standard InChI[11] - Reconnected layer (prefix: "
r"); contains the whole InChI of a structure with reconnected metal atoms; never included in standard InChI
The delimiter-prefix format has the advantage that a user can easily use a wildcard search to find identifiers that match only in certain layers.
| Structural formula | standard InChI |
|---|---|
InChI=1S/C2H6O/c1-2-3/h3H,2H2,1H3
| |
 |
InChI=1S/C6H8O6/c7-1-2(8)5-3(9)4(10)6(11)12-5/h2,5,7-10H,1H2/t2-,5+/m0/s1
|
InChIKey
[edit]The condensed, 27 character InChIKey is a hashed version of the full InChI (using the SHA-256 algorithm), designed to allow for easy web searches of chemical compounds.[6] The standard InChIKey is the hashed counterpart of standard InChI. Most chemical structures on the Web up to 2007 have been represented as GIF files, which are not searchable for chemical content. The full InChI turned out to be too lengthy for easy searching, and therefore the InChIKey was developed. There is a very small, but nonzero chance of two different molecules having the same InChIKey, but the probability for duplication of only the first 14 characters has been estimated as only one duplication in 75 databases each containing one billion unique structures. With all databases currently having below 50 million structures, such duplication appears unlikely at present. A recent study more extensively studies the collision rate finding that the experimental collision rate is in agreement with the theoretical expectations.[12]
The InChIKey currently consists of three parts separated by hyphens, of 14, 10 and one character(s), respectively, like XXXXXXXXXXXXXX-YYYYYYYYFV-P. The first 14 characters result from a SHA-256 hash of the connectivity information (the main layer and /q sublayer of the charge layer) of the InChI. The second part consists of 10 characters resulting from a hash of the remaining layers of the InChI, a single character indicating the kind of InChIKey (S for standard and N for nonstandard), and a character indicating the version of InChI used (currently A for version 1). Finally, the single character at the end indicates the protonation of the core parent structure, corresponding to the /p sublayer of the charge layer (N for no protonation, O, P, ... if protons should be added and M, L, ... if they should be removed.)[13][5]
Example
[edit]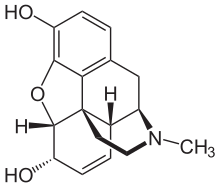
Morphine has the structure shown on the right. The standard InChI for morphine is InChI=1S/C17H19NO3/c1-18-7-6-17-10-3-5-13(20)16(17)21-15-12(19)4-2-9(14(15)17)8-11(10)18/h2-5,10-11,13,16,19-20H,6-8H2,1H3/t10-,11+,13-,16-,17-/m0/s1 and the standard InChIKey for morphine is BQJCRHHNABKAKU-KBQPJGBKSA-N.[14]
InChI resolvers
[edit]As the InChI cannot be reconstructed from the InChIKey, an InChIKey always needs to be linked to the original InChI to get back to the original structure. InChI Resolvers act as a lookup service to make these links, and prototype services are available from National Cancer Institute, the UniChem service at the European Bioinformatics Institute, and PubChem. ChemSpider has had a resolver until July 2015 when it was decommissioned.[15]
Name
[edit]The format was originally called IChI (IUPAC Chemical Identifier), then renamed in July 2004 to INChI (IUPAC-NIST Chemical Identifier), and renamed again in November 2004 to InChI (IUPAC International Chemical Identifier), a trademark of IUPAC.
Continuing development
[edit]Scientific direction of the InChI standard is carried out by the IUPAC Division VIII Subcommittee, and funding of subgroups investigating and defining the expansion of the standard is carried out by both IUPAC and the InChI Trust. The InChI Trust funds the development, testing and documentation of the InChI. Current extensions are being defined to handle polymers and mixtures, Markush structures, reactions[16] and organometallics, and once accepted by the Division VIII Subcommittee will be added to the algorithm.
Software
[edit]The InChI Trust has developed software to generate the InChI, InChIKey and other identifiers. The release history of this software follows.[17]
| Software and version | Date | License | Comments |
|---|---|---|---|
| InChI v. 1 | April 2005 | ||
| InChI v. 1.01 | August 2006 | ||
| InChI v. 1.02beta | Sep. 2007 | LGPL 2.1 | Adds InChIKey functionality. |
| InChI v. 1.02 | Jan. 2009 | LGPL 2.1 | Changed format for InChIKey. Introduces standard InChI. |
| InChI v. 1.03 | June 2010 | LGPL 2.1 | |
| InChI v. 1.03 source code docs | March 2011 | ||
| InChI v. 1.04 | Sep. 2011 | IUPAC/InChI Trust InChI Licence 1.0 | New license. Support for elements 105-112 added. CML support removed. |
| InChI v. 1.05 | Jan. 2017 | IUPAC/InChI Trust InChI Licence 1.0 | Support for elements 113-118 added. Experimental polymer support. Experimental large molecule support. |
| RInChI v. 1.00 | March 2017 | IUPAC/InChI Trust InChI Licence 1.0, and BSD-style | Computes reaction InChIs.[16] |
| InChI v. 1.06 | Dec. 2020 | IUPAC/InChI Trust InChI Licence 1.0[10] | Revised polymer support. |
| InChI v. 1.07.1 | Aug. 2024 | MIT License | Code moved to GitHub |
Adoption
[edit]The InChI has been adopted by many larger and smaller databases, including ChemSpider, ChEMBL, Golm Metabolome Database, and PubChem.[18] However, the adoption is not straightforward, and many databases show a discrepancy between the chemical structures and the InChI they contain, which is a problem for linking databases.[19]
See also
[edit]- Molecular Query Language
- Simplified molecular-input line-entry system (SMILES)
- Molecule editor
- SYBYL Line Notation
- Bioclipse generates InChI and InChIKeys for drawn structures or opened files
- the Chemistry Development Kit uses JNI-InChI to generate InChIs, can convert InChIs into structures, and generate tautomers based on the InChI algorithms
Notes and references
[edit]- ^ "IUPAC International Chemical Identifier Project Page". IUPAC. Archived from the original on 27 May 2012. Retrieved 2012-12-05.
- ^ Heller, S.; McNaught, A.; Stein, S.; Tchekhovskoi, D.; Pletnev, I. (2013). "InChI - the worldwide chemical structure identifier standard". Journal of Cheminformatics. 5 (1): 7. doi:10.1186/1758-2946-5-7. PMC 3599061. PMID 23343401.
- ^ "What on Earth is InChI?". IUPAC 100. Retrieved 10 May 2024.
- ^ "The InChI Trust and IUPAC". InChI Trust. Retrieved August 22, 2022.
- ^ a b c d e Heller, S.R.; McNaught, A.; Pletnev, I.; Stein, S.; Tchekhovskoi, D. (2015). "InChI, the IUPAC International Chemical Identifier". Journal of Cheminformatics. 7: 23. doi:10.1186/s13321-015-0068-4. PMC 4486400. PMID 26136848.
- ^ a b "The IUPAC International Chemical Identifier (InChI)". IUPAC. 5 September 2007. Archived from the original on October 30, 2007. Retrieved 2007-09-18.
- ^ E.L. Willighagen (17 September 2011). "InChIKey collision: the DIY copy/pastables". Retrieved 2012-11-06.
- ^ Goodman, Jonathan M.; Pletnev, Igor; Thiessen, Paul; Bolton, Evan; Heller, Stephen R. (December 2021). "InChI version 1.06: now more than 99.99% reliable". Journal of Cheminformatics. 13 (1): 40. doi:10.1186/s13321-021-00517-z. PMC 8147039. PMID 34030732.
- ^ McNaught, Alan (2006). "The IUPAC International Chemical Identifier:InChl". Chemistry International. Vol. 28, no. 6. IUPAC. Retrieved 2007-09-18.
- ^ a b "IUPAC/InChI-Trust Licence for the International Chemical Identifier (InChI) Software" (PDF). IUPAC/InChI-Trust. 2020. Retrieved 2022-08-09.
- ^ a b c Heller, Stephen R.; McNaught, Alan; Pletnev, Igor; Stein, Stephen; Tchekhovskoi, Dmitrii (2015). "InChI, the IUPAC International Chemical Identifier". Journal of Cheminformatics. 7: 23. doi:10.1186/s13321-015-0068-4. PMC 4486400. PMID 26136848.
- ^ Pletnev, I.; Erin, A.; McNaught, A.; Blinov, K.; Tchekhovskoi, D.; Heller, S. (2012). "InChIKey collision resistance: An experimental testing". Journal of Cheminformatics. 4 (1): 39. doi:10.1186/1758-2946-4-39. PMC 3558395. PMID 23256896.
- ^ "Technical FAQ - InChI Trust". inchi-trust.org. Retrieved 2021-01-08.
- ^ "InChI=1/C17H19NO3/c1-18..." Chemspider. Retrieved 2007-09-18.
- ^ InChI Resolver, 27 July 2015
- ^ a b Grethe, Guenter; Blanke, Gerd; Kraut, Hans; Goodman, Jonathan M. (9 May 2018). "International chemical identifier for reactions (RInChI)". Journal of Cheminformatics. 10 (1): 45. doi:10.1186/s13321-018-0277-8. PMC 4015173. PMID 24152584.
- ^ Downloads of InChI Software, accessed Jan. 8, 2021.
- ^ Warr, W.A. (2015). "Many InChIs and quite some feat". Journal of Computer-Aided Molecular Design. 29 (8): 681–694. Bibcode:2015JCAMD..29..681W. doi:10.1007/s10822-015-9854-3. PMID 26081259. S2CID 31786997.
- ^ Akhondi, S. A.; Kors, J. A.; Muresan, S. (2012). "Consistency of systematic chemical identifiers within and between small-molecule databases". Journal of Cheminformatics. 4 (1): 35. doi:10.1186/1758-2946-4-35. PMC 3539895. PMID 23237381.
External links
[edit] InChI (P234) (see uses)
InChI (P234) (see uses)
- IUPAC InChI site
- Description of the canonicalization algorithm
- Googling for InChIs a presentation to the W3C.
- InChI Release 1.02 InChI final version 1.02 and explanation of Standard InChI, January 2009
- NCI/CADD Chemical Identifier Resolver Generates and resolves InChI/InChIKeys and many other chemical identifiers
- PubChem online molecule editor that supports SMILES/SMARTS and InChI
- ChemSpider Compound APIs ChemSpider REST API that allows generation of InChI and conversion of InChI to structure (also SMILES and generation of other properties)
- MarvinSketch from ChemAxon, implementation to draw structures (or open other file formats) and output to InChI file format
- BKchem implements its own InChI parser and uses the IUPAC implementation to generate InChI strings
- CompoundSearch implements an InChI and InChI Key search of spectral libraries
- SpectraBase implements an InChI and InChI Key search of spectral libraries
- JSME Archived 2015-01-06 at the Wayback Machine is a free JavaScript based molecular editor that generates InChI and InChI Key in a web browser, which allows for easy web searches of chemical compounds

